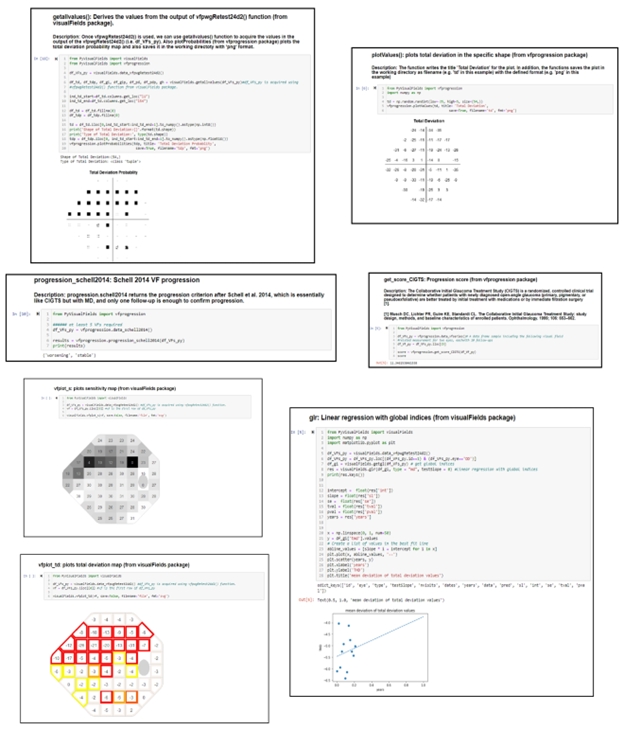
AMD is characterized by 4 distinct stages depending on physiological characteristics. Intermediate AMD (iAMD) is a critical stage for many individuals as it can be characterized by significant retinal changes, initial visual deficits, and is a risk factor for progression to the advanced forms: neovascular (“wet”) and geographic atrophy (“dry”).
Current clinical practice has established best-corrected visual acuity (BCVA) as the primary functional endpoint for AMD. However, recent research has noted that visual acuity may not be significantly altered by AMD progression, but contrast sensitivity may be a better metric for assessing functional deficits. (Vingopoulos et al 2021, Mones and Rubin) As such, a new functional tool called the quantitative contrast sensitivity function (qCSF) test has gained interest in monitoring AMD progression as it is able to differentiate between AMD staging based on its clinical results. With the help of AI, the test uses a Bayesian machine-learning model to adapt letter sizing (spatial frequency) and letter contrast depending on the patient’s correctness. Once the test is completed, qCSF provides critical metrics, such as contrast acuity (CA), that provide insight into the individual’s visual deficits, longitudinal progression, and retinal changes.
Specifically, our study reveals that certain biomarkers of iAMD, including subretinal drusenoid deposits and hyperreflective foci, are associated with changes in qCSF performance. Moreover, our results identified risk factors for AMD progression in both retinal biomarkers and qCSF metrics, further demonstrating that the test may prove useful in determining if progression is likely. This study helps to establish the importance of the qCSF test and its AI tools in monitoring AMD progression and its impact on visual function.
Ophthalmology Clinical Data Science Research Group Research Areas
Ophthalmology Clinical Data Science Research Group Research Areas
The Ophthalmology Clinical Data Science Research Group primarily focuses on leveraging extensive clinical databases, such as the IRIS® (Intelligent Research in Sight) Registry and internal patient data within the Massachusetts General Brigham system. Our research aims to develop and refine robust methodology to extract meaningful insights from these vast clinical registries. Through these efforts, we strive to enhance the understanding of ophthalmic conditions and treatment strategies across a variety of specialties in ophthalmology. Our lab’s publications cover a wide range of areas in ophthalmology, some of which include: the epidemiology of corneal conditions, risk factors for glaucoma and surgical complications, and treatment trends for retinal diseases.
Characterization of Acute Retinal Necrosis and Subsequent Retinal Detachment Risk: An IRIS® Registry Analysis
Systemic antiviral medications are typically part of the treatment regimen for all cases of acute retinal necrosis (ARN). Clinicians often add intravitreal antiviral medication injections and sometimes consider adjunctive early pars plana vitrectomy (PPV). Existing studies that have compared different treatment approaches for ARN have had modest sample sizes, given the rarity of this disease, and have reported conflicting results. This study aimed to determine the associations between initial treatment regimen for ARN and rate of retinal detachment (RD) and visual acuity outcomes at 6 and 12 months in a large national database.
Genetics of Diabetic Retinopathy
Pharmacogenetics
Risk Factors for Uveitis from Large Health Care Claim Databases
The risk factors for uveitis are not well understood. Funded through an NEI R21 grant and in collaboration with Dr. Brian VanderBeek at the University of Pennsylvania, Dr. Sobrin has led studies in a national healthcare claims database to the identify the risk of uveitis with various medications, including female hormonal therapy and statin therapy which both significantly impacted the risk of uveitis. We also investigated the utilization of osteoporosis monitoring in patients with uveitis who are treated with oral steroids in this large healthcare claims database.
Dr. Sobrin’s lab has been investigating the role of Vitamin D in the incidence of uveitis for several years in both our own hospital databases as well as in a national healthcare claims database, and these studies have consistently shown an association between low Vitamin D blood levels and increased risk of uveitis. In addition, Dr. Sobrin led a Mendelian randomization genetic study that provided support that low Vitamin D may be on the causal pathway to increased uveitis risk. This study leveraged our large Mass General Brigham Clinical and Genetic Repositories.
Non-Mydriatic Fundus-on-Phone 10 for glaucoma screening
The Friedman Lab, in collaboration with Aravind Eye Hospital in India, is evaluating the Non-Mydriatic Fundus-on-Phone 10 (NM-FOP 10) for glaucoma screening to explores sustainable screening options for regions with limited access to eye care.
Glaucoma is the second leading cause of blindness worldwide, affecting nearly 4 million people with vision-impairing glaucoma. In less developed countries, nearly 90% of those with glaucoma are unaware they have the disease. Timely detection prevents vision loss, but identifying high-risk patients in large, diverse populations remains challenging. Traditional methods rely on comprehensive eye exams by ophthalmologists, which are subjective, time-consuming and resource-intensive, particularly in areas with limited access to specialists.
The NM-FOP 10, developed by Remidio, offers an innovative approach to glaucoma screening. This non-mydriatic fundus camera captures high-resolution images of the optic nerve and assigns a glaucoma risk score for each patient without the need for pupil dilation. Although the device has been used in outpatient and glaucoma-specific clinics, this study is the first to assess its performance in a comprehensive ophthalmology clinic setting.
Dr. Friedman is collaborating with Aravind Eye Hospital in Pondicherry, India—the largest eye healthcare system in the world—to evaluate how effectively the NM-FOP 10 identifies patients who need referral to specialized glaucoma clinics compared to the current standard of care provided by comprehensive ophthalmologists. By assessing the device’s accuracy and reliability, the study aims to determine whether it can serve as an effective tool for glaucoma screening in broader clinical settings, ensuring that no patient in need of further examination is overlooked. Through this evaluation, the lab aims to develop a sustainable strategy to enhance access to timely care, improve early detection rates, and reduce the burden on ophthalmology clinics.
Retina Biobank
The Retina Biobank aims to collect and store biological samples, primarily from individuals with retinal diseases, as a source of big data for the research of genetics, proteomics, and metabolomics of these conditions. By analyzing DNA, serum, and other components, the project seeks to better understand the mechanisms of retinal diseases, contributing to improvements in diagnosis, treatment, and prevention. The biobank also serves as a valuable resource for broader research into various diseases.
Central Serous Chorioretinopathy
The Central Serous Chorioretinopathy (CSCR) study aims to identify genetic markers associated with the risk of developing CSCR. Patients’ blood is analyzed for genetic markers and compared to control groups. This study, funded by the National Institutes of Health (NIH), seeks to advance the understanding of genetic links to CSCR, which may eventually help improve diagnosis and treatment. Since CSCR is such a rare condition, these samples will serve as a repository for future adjacent research studies.
Structure-Based Network Analysis of Inherited Retinal Diseases
The project under Dr. Blake Hauser (MD, PhD), focusing on inherited retinal diseases, utilizes structure-based network analysis to predict the pathogenicity of genetic variants in proteins. The research integrates computational protein modeling to analyze changes in amino acid interactions, helping to understand how variants affect protein structure. We aim to map the gains and losses in protein interactions, especially at the molecular level, by analyzing the energetics of amino acid pairs. This approach enhances the understanding of variant impacts, supporting the identification of disease-associated mutations.
IRIS® Registry
Data Availability and Missingness in the IRIS® Database
Epidemiology of Diabetic Retinopathy
Improving Mass Screening for Diabetic Retinopathy Using Artificial Intelligence
The Friedman Lab is dedicated to improving mass screening for diabetic retinopathy (DR)—the leading cause of preventable blindness among American working-age adults—using artificial intelligence (AI).
Given that approximately 7.7 million Americans aged 40 and older are living with diabetic retinopathy (DR), the demand for effective and accessible screening is immense. Traditional screening methods, which rely on primary care physicians referring patients for annual dilated eye exams, face significant barriers including limited access to eye care, a shortage of specialists, and high costs. These barriers contribute to low compliance rates, with some health systems reporting adherence as low as 18-60%.
To address these challenges, the Friedman Lab is implementing a large-scale screening program across multiple community health locations, utilizing FDA-approved AI-enabled cameras (LumineticsCore, Digital Diagnostics) that automatically grade fundus images for signs of DR. This AI-driven approach simplifies and enhances the screening process, making it more accessible and equitable.
The lab’s research aims to understand and address the factors contributing to non-adherence to follow-up care after a positive DR screening, a critical gap in current healthcare practices. By identifying these barriers, Friedman Lab hopes to improve patient adherence to recommended ophthalmic care, ultimately preventing vision loss and improving eye health outcomes on a broad scale.
The Clinical Implications of Polygenic Risk Scores in Predicting Primary Open-angle Glaucoma
The Friedman Lab is investigating the clinical implications of polygenic risk scores (PRS) in predicting primary open-angle glaucoma (POAG) across diverse populations.
Glaucoma can lead to irreversible vision loss if not diagnosed and treated promptly, making timely diagnosis and continuous monitoring crucial for preserving vision. Traditionally, glaucoma has been diagnosed through screening and ongoing ophthalmic examinations. However, these methods can sometimes miss cases, as glaucoma patients may remain asymptomatic for years.
Polygenic risk scores (PRS) offer a promising alternative. By providing a clinically useful measure of aggregate genetic burden, PRS enables individual risk scoring and may facilitate earlier detection and continuous monitoring for those at risk of developing primary open-angle glaucoma (POAG). With 127 identified risk loci, POAG is known for its strong hereditary component.
The PRS study aims to develop and validate a novel POAG PRS to determine if individuals with higher genetic risk scores are more likely to develop glaucoma. Using data from a multi-ethnic genome-wide association study, the research incorporates cross-ancestry effect estimates and advanced software algorithms to evaluate the PRS’s effectiveness in predicting glaucoma risk across various biobank populations, including the UK Biobank and Massachusetts General Hospital-based cohorts such as the BioMe BioBank and Partners HealthCare Biobank. Additionally, the project investigates how environmental exposures impact glaucoma risk by integrating PRS data with responses from environmental exposure questionnaires.
Dr. Friedman is a co-investigator in this initiative, and Janey Wiggs is the principle investigator. This approach aims to enhance understanding of genetic risk factors in glaucoma and their interaction with environmental influences, potentially leading to more personalized and comprehensive diagnostic and treatment strategies.
Real-world Implementation of Ophthalmic Telemedicine, Digital Health
Dr. Armstrong’s research focuses on the real-world implementation of ophthalmic telemedicine, digital health, and artificial intelligence to enhance the screening, triage, and diagnosis of ocular conditions. By leveraging these technologies, Dr. Armstrong’s lab aims to improve access to care in underserved communities and reduce health disparities to develop scalable, innovative solutions that ensure equitable and efficient delivery of ophthalmic care.

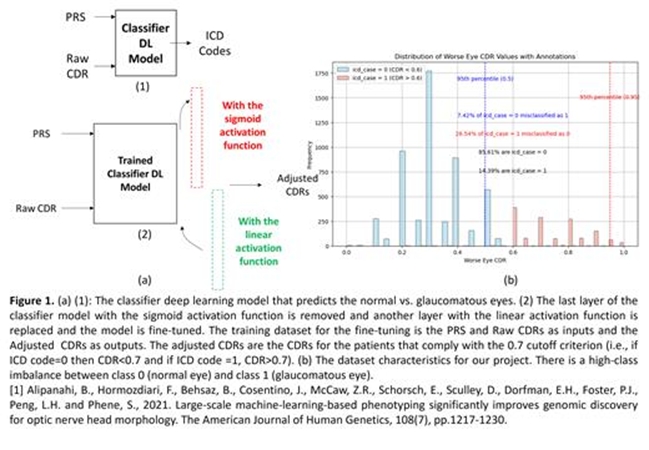
Genetically Adjusted Optic Cup to Disc Ratio (CDR) Using a Two-Phase Training Deep Learning Model
Genetically Adjusted Optic Cup to Disc Ratio (CDR) Using a Two-Phase Training Deep Learning Model
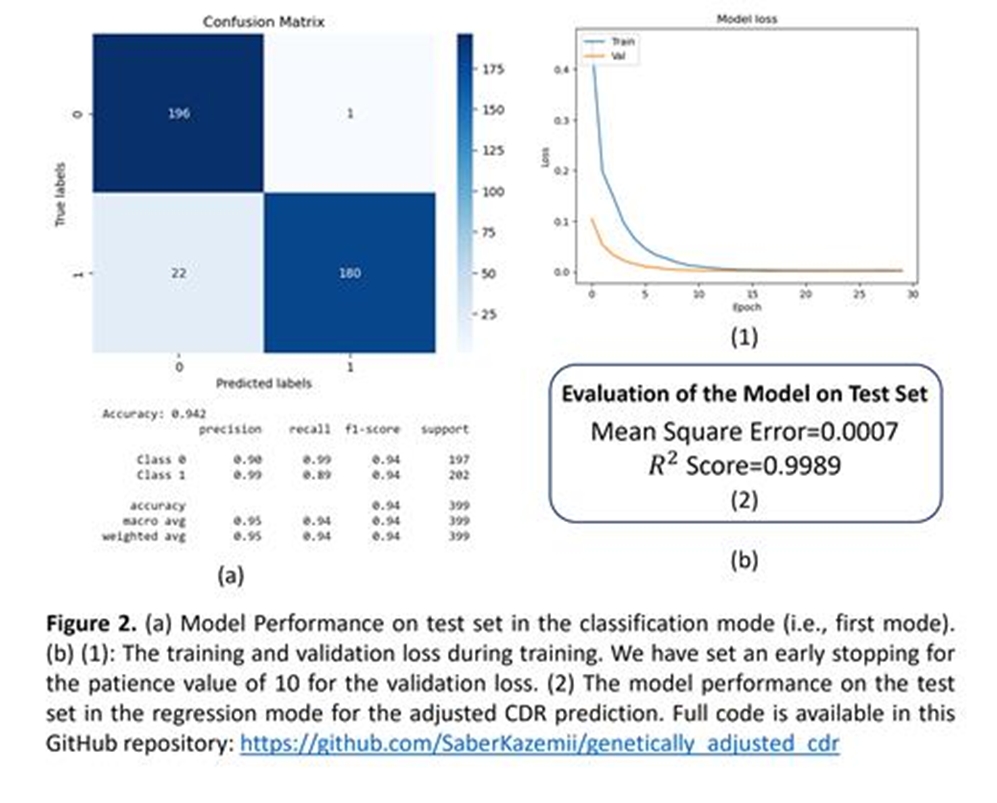
This project aims to enhance the diagnostic accuracy of glaucoma by refining the optic cup-to-disc ratio (CDR) assessment using genetic data. While a CDR cut-off of 0.7 is commonly used to indicate glaucoma, it fails to account for many clinical cases where patients with glaucoma present with a CDR less than 0.7, and vice versa. To address this, we calculated a CDR polygenic risk score (PRS) for 6575 patients from the Mass General Brigham (MGB) clinical biobank, based on loci associated with primary open-angle glaucoma (POAG) from a large genome-wide association study. We developed a deep learning model with a two-phase training approach: initially classifying patients’ glaucoma status using PRS and raw CDR values, then adjusting the CDR based on genetic risk. Our cohort included 53% females, predominantly of European ancestry, with a significant class imbalance favoring non-glaucomatous eyes. The model was validated against 399 manually reviewed samples, achieving a high classification accuracy with F1 scores of 0.95 and 0.94 for non-glaucomatous and glaucomatous eyes respectively, and an R2 score of 0.9989 in regression tests. This approach demonstrated improved clinical accuracy in CDR assessment, offering a more reliable diagnostic tool for glaucoma.
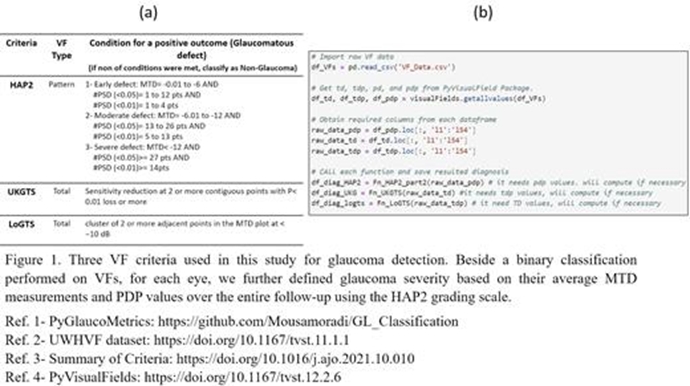


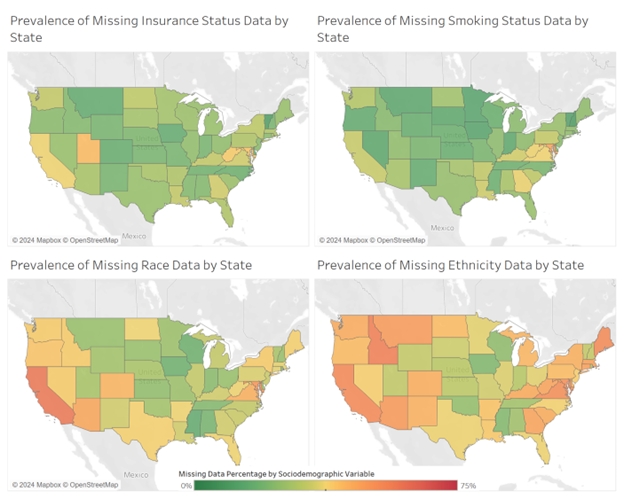
PyGlaucoMetrics: An Open-Source Multi-Criteria Glaucoma Defect Evaluation
This project aims to enhance the measurement of glaucomatous vision loss and address the challenge of unreliable ICD codes in clinical data by developing an open-source Python repository that standardizes and assesses Visual Field (VF) data. The repository automates the identification of glaucoma signs based on predefined criteria. Our Python toolbox categorizes glaucomatous VF defects using three established criteria: Hodapp-Anderson-Parrish 2 (HAP2), United Kingdom Glaucoma Treatment Study criteria (UKGTS), and Low-pressure Glaucoma Treatment Study criteria (LoGTS). Analyses were performed on a publicly available dataset of Humphrey field analyzer (HFA) static perimetry from 7248 unique eyes (3871 patients). Preprocessing computes mean total deviation (MTD), total deviation probability (TDP), and pattern deviation probability (PDP) using the PyVisualFields library. Statistical analysis measured variance and reported the Intraclass Correlation Coefficient (ICC) among the criteria, revealing a high ICC of 0.988, indicating strong agreement. HAP2 criteria classified VFs into four stages: non-glaucomatous, early defect, moderate defect, and severe defect. Our results showed that HAP2, UKGTS, and LoGTS identified 17015, 17664, and 17090 VFs as glaucomatous, respectively, with HAP2 distinguishing 41% of eyes as non-glaucomatous, and the remaining classified as early, moderate, and severe glaucoma. Our GitHub repository offers a robust tool for glaucoma detection and VF defect staging, providing valuable insights for clinical applications and future research, and will be maintained and updated to ensure state-of-the-art capabilities.
An unsupervised deep learning method for identifying glaucoma progression patterns using longitudinal ganglion cell complex (GCC) scans
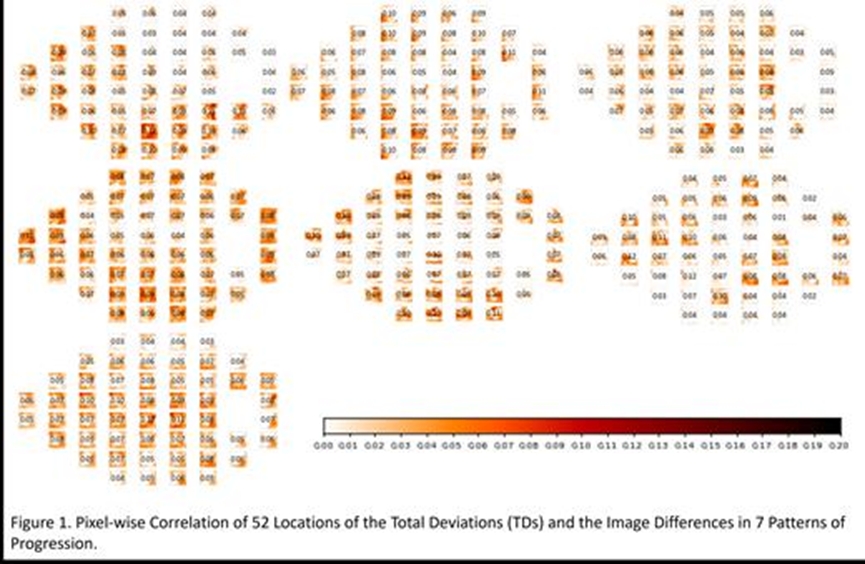
This study utilizes unsupervised deep learning to identify distinct glaucoma progression patterns from macular ganglion cell complex (GCC) scans and examines their relationship with visual field (VF) metrics and clinical parameters. We analyzed 28,574 OCT scans from 5,031 patients, each having at least one ICD code for glaucoma (H40.xx). Longitudinal pairs of GCC scans from different visits were registered using the Nifty registration method to standardize camera angles and magnification. We calculated pixel-wise difference maps for these scan pairs and used them to train an autoencoder. The number of clusters was determined using AIC and BIC scores, the elbow point, and the silhouette score, and clustering was performed using a Gaussian Mixture Model (GMM). We assessed the correlation between these progression patterns and VF total deviations (TDs), intraocular pressure (IOP), and cup-to-disc ratios (CDRs). The cohort comprised 58% females, 11.31% Hispanic, 79.27% White, 13.68% Black, 6.68% Asian, and less than 1% from other races, with an average time difference of 1.16 years between visits. We identified seven optimal OCT progression patterns showing strong regional correlations with VF TDs and significant correlations with IOP, CDR changes, and VF mean deviations (MDs). This study demonstrates the effectiveness of an unsupervised AI approach for detecting OCT-based glaucoma progression patterns and highlights regions associated with key clinical parameters, potentially enabling earlier identification of glaucoma progression using OCT scans.
A graph neural network-based clustering method for glaucoma detection from OCT scans considering uncertainties in the number of clusters
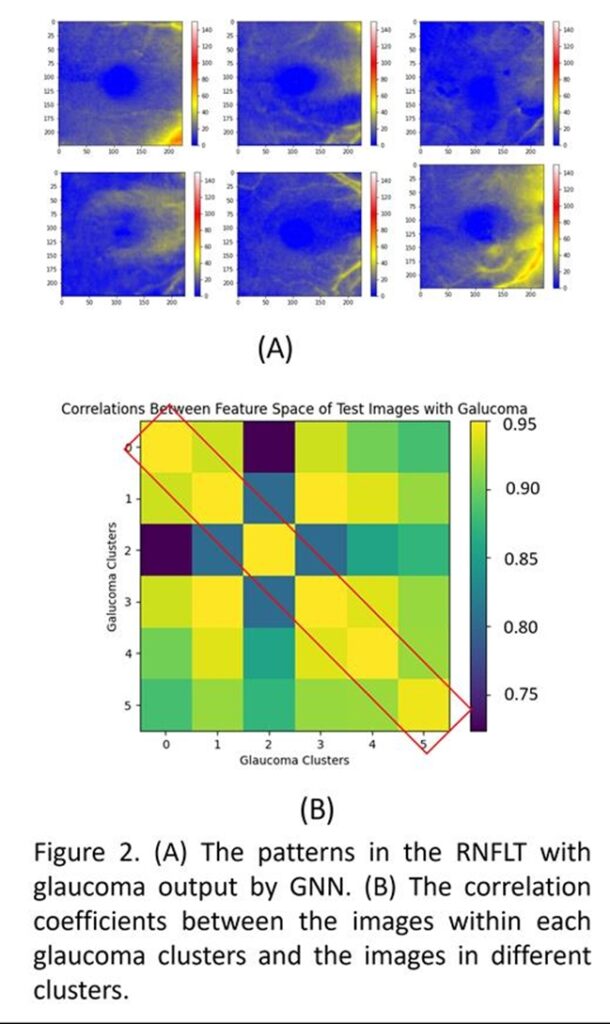
This project aims to enhance downstream tasks such as glaucoma detection and genetic analysis by clustering unlabeled OCT scans using an artificial intelligence model based on a graph neural network (GNN) that automatically determines the number of clusters and organizes the images. We first extracted the mean deviation (MD) from visual tests conducted within six months before and after acquiring retinal nerve fiber layer thickness (RNFLT) images from the Mass Eye and Ear dataset. Images were labeled as normal (MD ≥ 0 dB) or non-normal (MD < 0 dB). An encoder trained through unsupervised representation learning was used to extract feature spaces from the images. A GNN constructed using K-nearest neighbor (K=10) was trained with the feature space of the labeled images, where each image represented a node and its feature space represented the node feature. The graph was iteratively reconstructed based on the labels until it converged into a final graph. During inference, the GNN clustered the unlabeled feature space into an unknown number of clusters. The model was trained on 18,507 scans from 5,559 patients and tested on an unforeseen dataset of 5,053 scans from 2,064 patients. The GNN produced 19 clusters for all scans, and after filtering out the normal clusters, 6 clusters (patterns) remained for non-normal images. The Pearson’s correlation coefficient showed a strong correlation among non-normal eyes, with even stronger correlations within individual clusters. This proof of concept addresses the challenge of identifying the number of patterns in unlabeled OCT scans, facilitating glaucoma detection and genetic analysis, such as genome-wide association studies (GWAS). Moreover, the hierarchical learning of the graph architecture allows clustering of OCT images taken from different devices using a single trained GNN.

Genome wide discovery via feature space mapping of deep-learning derived clinical OCT phenotypes to the UK biobank
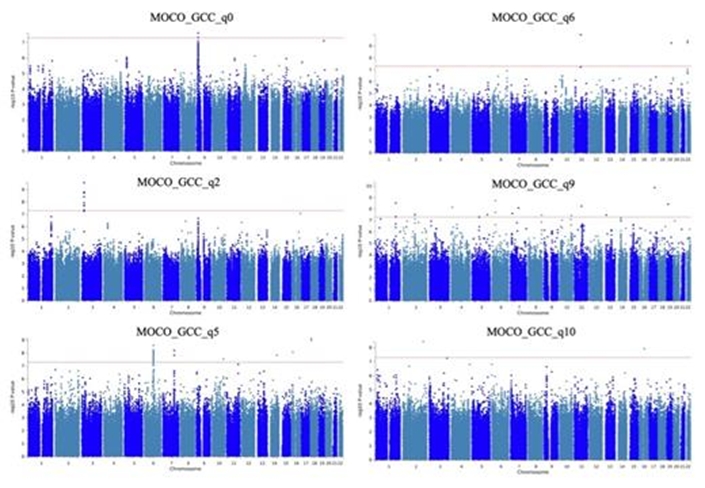
In this study, we leverage large population-based biobanks and a clinical glaucoma dataset to discover macular OCT-based phenotypes using unsupervised deep learning (DL) and test these phenotypes in the UK Biobank for genome-wide discovery. We used 7,966 high-quality macular optical coherence tomography (OCT) scans from 4,926 glaucoma patients in the Mass Eye and Ear (MEE) dataset to train a DL model employing unsupervised representation learning, data augmentation, and contrastive learning. The trained encoder associated a 3-channel image with a 128-element feature vector, which was then reduced in dimensionality using UMAP and clustered with a Gaussian Mixture Model. The optimal number of clusters was determined using probabilistic model selection. The trained DL model mapped 86,115 GCC thickness maps of 50,115 UK Biobank subjects into the feature space, followed by GWAS for each GMM-identified feature dimension using linear mixed models adjusted for age, sex, and ancestry.
We identified 11 optimal macular GCC patterns in the MEE dataset, which were successfully mapped to the UK Biobank, except for one pattern (q3). GWAS analysis, performed on 9,329,555 imputed genotypes after quality control, revealed no evidence of unaccounted stratification (lambdaGC 1.000-1.0208). Several independently associated SNPs (range: 1-15) were identified for each phenotype, with numerous genomic regions associated with at least one variant/phenotype linked to various biological processes, including glucocorticoid interaction, neurological development/disease, cholesterol synthesis, immune response, and malignancy. Additionally, the genomic regions were enriched for association with interferon-I activity, based on MAGMA analysis.
This study demonstrates that OCT-derived DL phenotypes discovered in a clinical cohort can be successfully transferred to an independent large biobank, enabling the potential discovery of disease-relevant genetic variants.

A Performance Evaluation Method for Unsupervised OCT Phenotype Discovery using Deep Learning
We have previously demonstrated the utility of deep unsupervised phenotype discovery with OCT imaging for genetic analysis. However, this method faces challenges in determining the optimal number of patterns and lacks ground truth for performance evaluation. In this study, we propose an unsupervised performance evaluation method that establishes a criterion for clustering performance evaluation, which can also determine the number of patterns.
We developed and trained two deep learning (DL) models: an autoencoder that learns features of OCT macular images in a pixel-wise manner, and an unsupervised representation learning-based method that maps OCT macular images to 128-element feature vectors using data augmentation and contrastive learning without human annotation. After dimensionality reduction with UMAP and probabilistic model selection (AIC/BIC), we defined the number of patterns for the dataset from the output embedding of the trained DL models. Clustering was performed using the Gaussian Mixture Model (GMM). Correlation analysis was then conducted for images within clusters and across different clusters in both pixel space and feature space.
We utilized 86,115 images from the UK Biobank (UKBB), 17,722 images from the LIFE dataset, and 8,436 images from the Mass Eye and Ear (MEE) dataset, including retinal nerve fiber layer (RNFL) and ganglion cell-inner plexiform layer (GCIPL) contour maps. While pixel space correlation analysis did not yield meaningful results, feature space correlation analysis demonstrated a stronger correlation between images within clusters compared to those in different clusters. The correlation in feature-wise clustering was higher than in pixel-wise clustering. Additionally, we observed that some patterns identified in one dataset were not present in another, indicating variability among datasets.
This study proposes a criterion for validating the optimal number of OCT-based patterns obtained using unsupervised deep learning. Importantly, we demonstrate that these phenotypes can be represented in external datasets, reinforcing their potential utility for future genetic discovery and risk prediction.


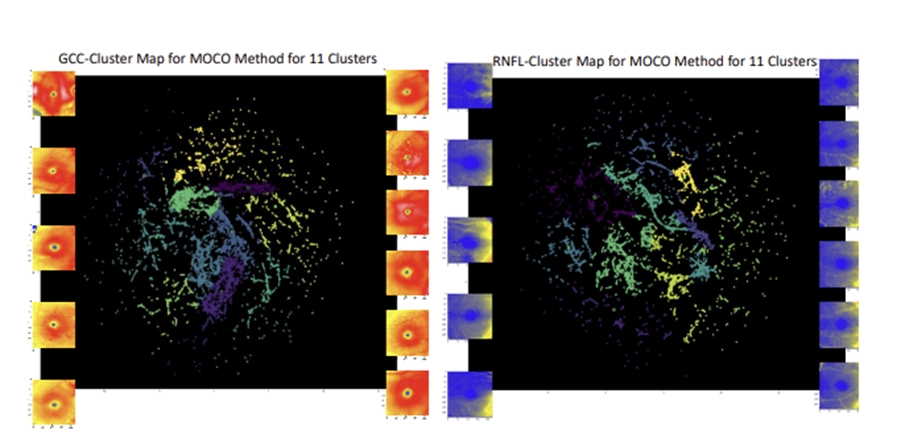


An artificial intelligence method for phenotyping of OCT scans using unsupervised and self-supervised deep learning
Artificial intelligence (AI) has been increasingly used to analyze optical coherence tomography (OCT) images to better understand physiology and genetic architecture of ophthalmic diseases. However, to date, research has been limited by the inability to transfer OCT phenotypes from one dataset to another. In this work, we propose a new AI method for phenotyping and clustering of OCT-derived retinal layer thicknesses using unsupervised and self-supervised methods in a large clinical dataset using glaucoma as a model disease and subsequently transfer our phenotypes to a large biobank. The model includes a deep learning model, manifold learning, and a Gaussian mixture model. We also propose a correlation analysis for the performance evaluation of our model based on Pearson correlation coefficients. Our model was able to identify clinically meaningful OCT phenotypes and successfully transfer phenotypes from one dataset to another. Overall, our results will contribute to stronger research methodologies for future research in OCT imaging biomarkers, augment testing of OCT phenotypes in multiple datasets, and ultimately improve our understanding of pathophysiology and genetic architecture of ocular diseases.
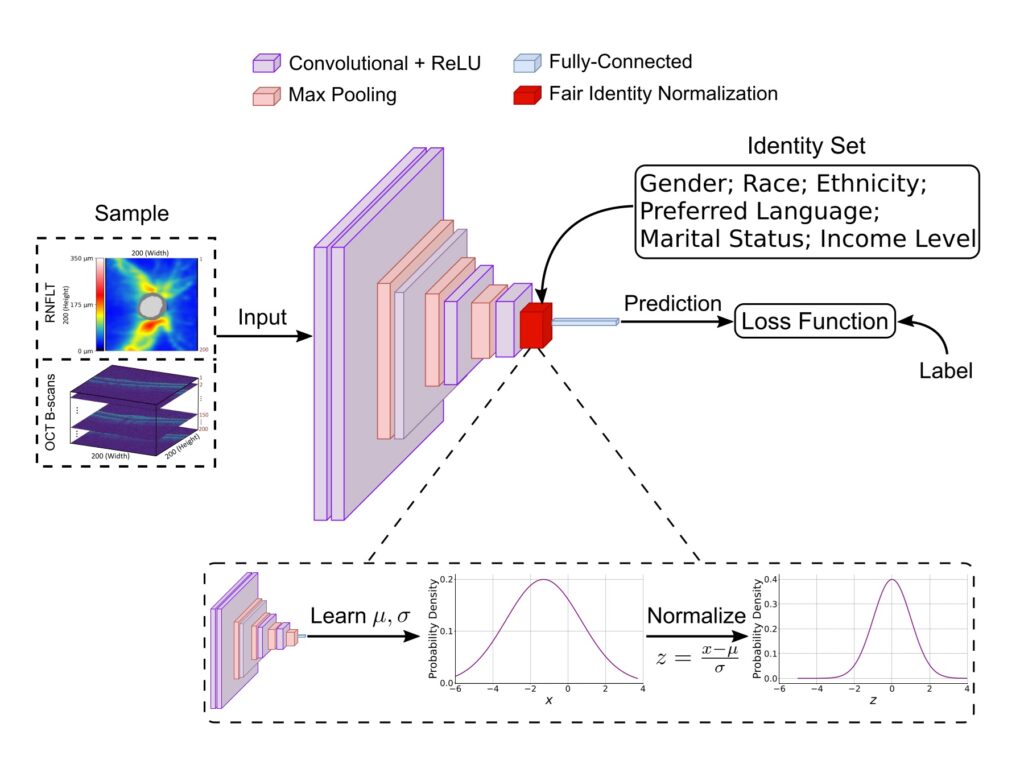
A significant number of human diseases disproportionately impact racial and ethnic minorities, as well as socioeconomically disadvantaged groups. This disparity largely stems from unhealthy lifestyle choices and inadequate healthcare access often associated with poverty. For example, vision loss due to glaucoma, a lead cause of irreversible blindness globally, is more severe in Blacks than in Whites and Asians. In addition, many diseases are underdiagnosed in racial and ethnic minorities and socioeconomically disadvantaged groups disproportionately impacted. For instance, it has been reported that 50% of people with glaucoma do not know they have the disease, and Blacks and Hispanics are 4.4 times and 2.5 times greater odds of having undiagnosed and untreated glaucoma than non-Hispanic Whites. Disease detection with deep learning using medical imaging data is promising to provide affordable disease screening to alleviate societal disease burden and reduce health disparities between different demographic identity groups, which can be deployed in primary care and pharmacies without needing the subjects to visit the more expensive and busy specialty clinics. Potential deep learning systems for automated disease screening should promote healthcare equality prior to clinical implementation.
Our team is dedicated to developing equitable AI models for disease screening with a special focus on ocular diseases. Recently, we have developed a fair identity normalization technique to equalize feature importance between different identity groups to improve model performance equity. Our team is committed not only to developing novel equitable AI techniques but also to releasing public datasets that can be used to facilitate a greater community of researchers to study medical AI equity. The success of this project will greatly benefit racial and ethnic minorities and socioeconomically disadvantaged groups with more equitable disease screening. Our equitable AI models developed based on the application of eye disease screening can be generalized to be used in other disease screening tasks to make a broader impact on various medical AI applications.
AI for identifying imaging endophenotypes for GWAS
Glaucoma is the first cause of blindness worldwide, which is a sophisticated multi-variable disease that engenders structural and functional variations. Glaucoma in terms of genetic analysis has been hindered due to the complicated structure of the retina layers and the limitation of the computation resources. The retina is a deeply complex structure of the eye that is responsible for receiving incoming light and providing some pre-processing and sending that information to the brain. It comprises several tissue layers that are involved in the process of the input light. In a type of glaucoma disease, any layers of the retina can be affected which makes it important to have a precise understanding of each layer in the retina. Optical Coherence Tomography (OCT) is an innovative and non-invasive imaging technique that plays an instrumental role in retinal diagnostics. It operates based on the principles of interferometry to capture high-resolution cross-sectional images, often termed ’optical biopsies’. OCT imaging provides us with the ability to clearly differentiate between various stratified layers of the retina. It achieves this by measuring the echo time delay and magnitude of backscattered light, thereby creating a detailed map of the retinal structure with 5-10 micrometers accuracy that is sufficient to identify subtle morphological changes in the retina which could be indicative of early-stage ocular diseases. Consequently, OCT has emerged as an invaluable tool in both the clinical and research contexts, contributing significantly to our understanding of the retina and its associated pathologies. Due to the complexity of the OCT images and the limitation of computational resources, precise analysis of the OCT images was still impossible. However, artificial intelligence (AI) has recently empowered researchers for more complex analysis in all areas and in fact, it has become an inseparable part of ophthalmology nowadays. The AI models help us to extract the features behind the data with high dimensionality like 3D medical images. Also, thanks to recent computational resources like graphical processing units (GPUs), more dense datasets can be analyzed with a better understanding of the feature variations in complex organs like the retina. However, the use of AI algorithms has been limited by the data annotation by experts [3] while large-scale biobank studies with multi-modal data have been started (e.g., UK biobank [4]). This is useful for discovering hidden structural patterns of OCT-derived macular thickness layers using deep learning (DL) among subjects with high polygenic risk (PRS) for primary open-angle glaucoma (POAG). Subsequently, we use these endophenotypes to discover novel genomic loci.
Our team proposes to uncover the hidden structural patterns of OCT-derived retina layers with two AI models. These patterns are useful for further genetic-wide analysis studies (GWAS). The proposed AI models then group semantically similar images. We also provide an unsupervised evaluation method that sets a ground truth for clustering evaluation and can be used for determining the number of clusters. We also investigate the derived endophenotypes for the longitudinal images to see if there is a correlation between the patterns and also the progress of glaucoma among the patients.

Vision loss detection and progression prediction
The long onset period and unnoticed symptoms of many ocular diseases make it hard for patients and clinical practitioners to be aware of the functional impairments. For instance, a high prevalence ophthalmic disease—glaucoma—often starts with small scotomas (blind spots) in the periphery, and is easy to be ignored. Glaucoma usually affects the peripheral area of the retina at the early stage, which may affect functional vision so subtly during the initial period that it goes unnoticed in the patient’s daily life often for over many years. However, once it turns to the advanced stage, the central visual acuity is affected severely, leading to irreversible functional visual loss, rapid decrease of living quality, and even the loss of eyeball. Similarly, diabetic retinopathy is another ocular disease that is asymptomatic during the initial stage but will heavily affect visual function during the proliferative stage. The prevalence of diabetes and prediabetes in the United States are 14.6% and 37.5%, respectively. And 34.6% of diabetic patients are estimated to have diabetic retinopathy but few of them are diagnosed and have a systemic clinical measurement. Diseases like glaucoma and diabetic retinopathy often go unnoticed until the moderate-to-advances stages of the disease. At the same time, many ocular diseases are irreversible, which makes early detection extremely important to timely initiate or adjust ocular therapy before diseases progress to stages that cause severe loss of living quality.
However, the diagnosis and detection of functional worsening of ocular disease are challenging, as measurements are noisy, and the differentiation between true disease progression and random fluctuations or measurement artifacts is difficult. Besides, the distinguishment between normal people and patients with ocular diseases is not that obvious in real life. People with ocular hypertension required long-term follow-up to prevent them to develop glaucoma. Meanwhile, people with normal-tension glaucoma are hard to be distinguished from normal people and need systemic detection to be found and diagnosed. How to develop an efficient detective method that meets both the screening and follow-up requirements of most ocular diseases is important and urgent.
Our team aims to better characterize the functional loss from ocular diseases like glaucoma with the goal of developing novel diagnostic methods and improving the prediction of how those diseases may progress in the future.
We developed and evaluated a novel scheme of representative patterns for glaucomatous vision loss based on unsupervised machine learning and explored the impact of individual anatomical parameters on it. These patterns help to distinguish true glaucomatous functional loss from the various and frequent measurement artifacts and from damages caused by other ocular diseases and contribute to the diagnosis of vision loss and its progression. Apart from the improvement of diagnostic and prognostic methods, our team also study the optimal scheduling of patients, i.e., the optimal time intervals for patient testing to maximize the information gain of each test.
The translational potential of this work related to diagnostic improvements is not only reflected by the publications, but also by the patents—namely one patent on modeling of glaucomatous visual field loss and another patent on an algorithm to determine the optimal scheduling intervals for patient testing.

Equitable AI for disease screening
Equitable AI for disease screening
AI for medical data cleaning
The advent of diagnostic methods such as optical coherence tomography (OCT) and standard automated perimetry has greatly enhanced the efficacy of identifying and monitoring ocular diseases. Although the data generated by these techniques offer valuable insights into retinal structural damage and functional vision loss, they are inherently susceptible to artifacts and noise. These factors can potentially lead to imprecise and unreliable diagnostic results.
For instance, clinicians commonly use circumpapillary RNFL thickness (RNFLT), obtained from spectral-domain OCT, as the principal structural metric for diagnosing and tracking glaucoma. However, widespread imaging artifacts often result from segmentation failures in scenarios of poor imaging quality or image defects, which frequently undermine the clinical value of RNFLT. Likewise, visual field (VF) testing, such as the 24-degree test, is a standard functional diagnostic measure for glaucoma. Yet, the impact of VF noise, causing significant variation in test-retest results, constrains its usefulness.
Moreover, the growing amount of noisy patient data presents a significant challenge to the viability and dependability of artificial intelligence (AI)-based diagnostic systems. These systems require training and evaluation on high-quality, clean medical data. It is indisputable that both clinical practice and AI system development should utilize clean, high-quality data, thereby creating a reliable and trusted ecosystem among practitioners, patients, and AI systems.
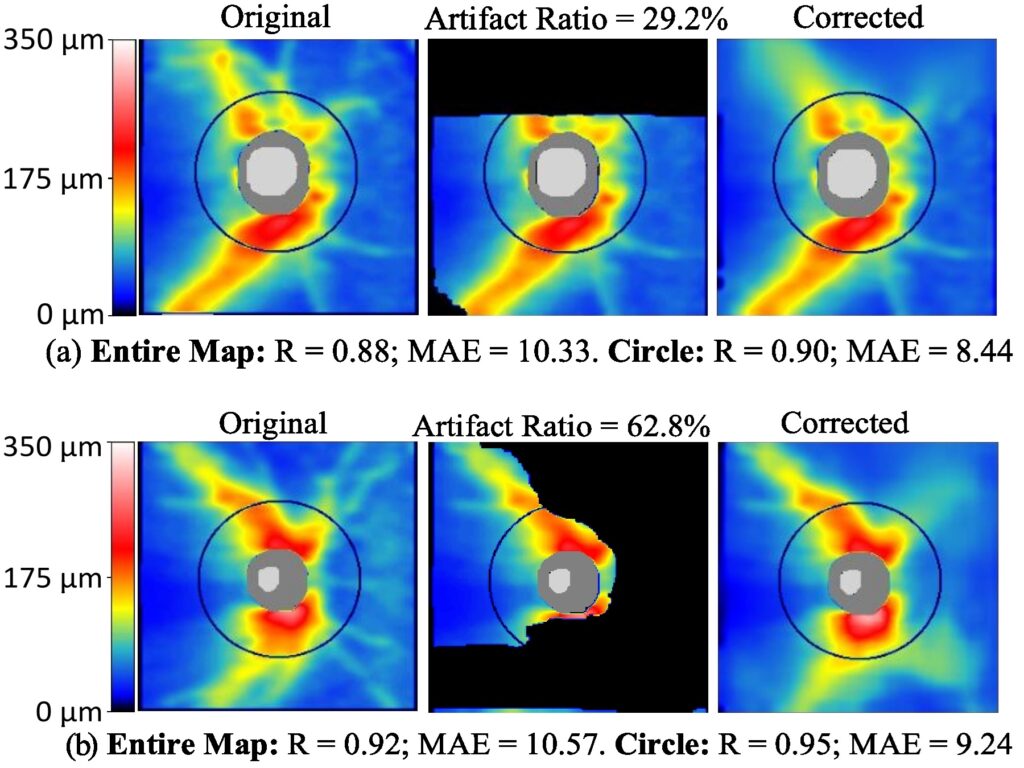
Our team is dedicated to developing AI-based data cleaning technologies, aiming to provide cleaner data and enhance diagnostic outcomes for eye diseases. For instance, we’ve developed an innovative deep learning framework called RNFLTCorrect to address OCT artifacts. This AI system was built and assessed using a substantial population of 24,257 glaucoma patients, encompassing 111,966 RNFLT maps. RNFLTCorrect has demonstrated high accuracy in artifact correction, enhancing VF prediction and progression forecasting in glaucoma. In addition, we’re developing AI methods to reduce noise in 24-degree VF data, which can significantly enhance the structure-function correlation. This progress has the potential to assist clinicians in diagnosing and monitoring glaucoma more effectively.

Interpretable AI for pathophysiology discovery
Retinal diseases are a significant global health concern affecting an estimated 2.2 billion individuals worldwide. Artificial intelligence (AI) and deep learning models are increasingly being utilized in the clinic within the field of ophthalmology. Over the last 5-year, most of the various AI diagnostic systems developed are based on fundus photography. Although fundus photography is one of the most common clinical imaging modalities in evaluating retinal abnormalities, however, it does not provide layer information of the retina. The high-resolution imaging capability of optical coherence tomography (OCT) allows for detailed visualization of the various layers of the retina, making it a valuable noninvasive diagnostic method widely used for diagnosing, characterizing, and monitoring ocular pathologies OCT’s sensitivity and specificity are comparable, if not superior, to those of fundus photography. Consequently, the application of artificial intelligence (AI) to OCT imaging holds great promise for screening and monitoring retinal diseases. Global and individual retinal layer thickness changes from OCT images have been observed in age-related macular degeneration (AMD), diabetic retinopathy (Dr), and glaucoma as well as systemic disorders.
Poor renal function as assessed by Cystatin C and estimated glomerular filtration rate was associated with a thinner circumpapillary retinal nerve fiber layer thickness (cpRNFLT). An adverse lipid profile (i.e., low high-density lipoprotein (HDL) cholesterol, as well as high total, high non-HDL, high low-density lipoprotein cholesterol. Further studies showed that a thinner RNFL is associated with an increased risk of dementia, including Alzheimer disease. Smoking and Alcohol have also been proven to cause a thinning of the retinal layers.
These studies underline how the retina is not only affected by eye-specific pathologies but could be a window to identify systemic disorders. Most of the underlying studies, however, employ a linear model to assess the correlation between retinal layer thickness and diseases, which may not accurately capture the potential non-linear nature of the relationship between retinal microstructure and ocular and systemic disorders. In contrast, deep learning is naturally capable to quantify both linear and non-linear relationships.
Our team aims to design a deep learning model to quantify the retinal layer importance and regional importance of each retinal layer in predicting various ocular and systematic diseases.
In particular, we exacted the retinal layer thickness maps for the 10 retinal layers segmented by the Heidelberg Engineering software including retinal nerve fiber layer (RNFL), ganglion cell layer (GCL), inner plexiform layer (IPL), inner nuclear layer (INL), outer plexiform layer (OPL), outer nuclear layer (ONL), myoid zone (MZ), ellipsoid zone (EZ) and outer-photoreceptor segment (OS) combined (EZ+OS), interdigitation zone (IZ) and retinal pigment epithelium (RPE). We used each of the 10 layers individually and 10 layers together to predict diabetes diagnosis. For each layer, we quantify the regional importance on predicting the eye and systemic diseases. We use the area under the receiver operating characteristic curve (AUC) to measure the layer importance and regional importance for each layer.
The new knowledge of the layer importance and regional importance for each layer can shed light on how retina is affected by different eye and systemic diseases distinctly, which can be potentially used to better preserve vision.

Relationship between retinal structure and visual function in eye diseases
Many ocular diseases manifest themselves on the retina before functional deficits develop. There would be a delay in recognizing the retinal damage if we only rely on the functional tests especially when the peripheral retinal is firstly damaged during the initial stage of diseases like glaucoma and pigmentary degeneration of the retina. The structural tests will provide detailed information at the early stage of diseases, which is essential to early diagnosis and progression follow-up.
Even in the presence of functional impairment, functional testing procedures like optometry can be time-consuming and exhausting for patients. The retinal structural tests like fundus imaging and optical coherence tomography (OCT), on the other hand, are convenient even for patients at an advanced age, and often take only seconds to a few minutes, which is much shorter than functional tests.
Meanwhile, optometry is a subjective test and can be easily affected by patients’ age, recognition level, attention, and even training times. In contrast, fundus imaging and OCT are objective, accurate, and quantitative.
It’s undeniable that both the functional and structural tests are indispensable for comprehensive clinical measurement, including screening, diagnosis, treatment, and follow-up, but it’s also a vital goal for researchers and clinical healthcare providers to combine the functional and structural tests in an efficient way, take advantages of each test, broaden the application among the population as well as save time and clinical resources.
Therefore, our team is investigating the relationship between damages of retinal structure and their precise effects on functional vision in ocular diseases. In particular, we study patient measurements from fundus images and OCT results, investing their association with specific effects on functional vision. We apply image processing and statistical learning methods to large sets of paired structure-function measurements of several ocular diseases (diabetic retinopathy, glaucoma, macular degeneration) to predict the details of functional vision loss from fundus photography or optical coherence tomography. From previous achievements, we studied the relationship between the three-dimensional (3D) optic nerve head (ONH) related structure and glaucoma and recently identified an important retinal biomarker specifically for central vision loss in glaucoma. Our research has high clinical relevance and can be potentially translated into clinical practice for better glaucoma diagnosis, monitoring, and treatment.
Epidemiology and demographic features related to eye diseases
The public health system is continually reminded of the challenges posed by the limited understanding of the diseases. It was recommended that every public health agency should collect, assemble, analyze, and make available information on the health of the community, including statistics on health status, community health needs, and epidemiologic and other studies of health problems. Ocular diseases management is a crucial part of the health care system as some ocular diseases are of high prevalence, like cataract and myopia, and will heavily affect the living quality of people. In addition, some are closely related to other systemic diseases like diabetic retinopathy and retinal vascular occlusion.
Regarding the lack of comprehensive pathologic mechanism knowledge and the complexity of proper detection, diagnosis, and treatment, population-based cohort studies provide a choice to investigate the interactions of genes, environment, society, and personal lifestyle with disease onset and progression. Large, comprehensive, and systemic population-based cohort studies are necessary and are the trend of future clinical studies. There are two examples of cohort studies.
The American Academy of Ophthalmology IRIS® Registry (Intelligent Research in Sight) is one of the large clinical ocular disease registries in the United States. It consists of data from more than 349 million patient visits of 60 million unique patients in the United States (as of September 1, 2020). The IRIS® Registry collects 5 eye care measures, 10 general prevention and patient safety measures, and 18 outcome measures from over 3,800 ophthalmic practices in the United States, and continually tracks patients’ history in the same specialty, including initial patient visit, intervention, and longitudinal follow-up. The IRIS platform provides a large-scale glimpse into the trends of eye disease, features, and real-world conditions such as patients’ identification and diagnosis, choices of treatment, and outcomes.
LIFE-Adult-Study is a population-based study conducted by the Leipzig Research Centre for Civilization Diseases (LIFE) in Leipzig, Germany. The study collects the baseline examination and follow-up results of 10,000 participants that were randomly selected from 550,000 inhabitants in Leipzig. The baseline examination was finished before November 2014, and the follow-up examination commenced in 2016 and is running till now. The measurements are not limited to ophthalmic measurements like high-quality non-mydriatic fundus imaging and optical coherence tomography (OCT) results, but also comprise structured personal interviews, self-administered questionnaires, physical and medical examinations, psychometric tests, more than 80 clinical chemical biomarkers as well as comprehensive molecular-genetic profiling comprising genotyping, transcriptome, metabolome, and proteome analyses based on blood and urine samples. Some participants have additional cognition and depression tests and an MRI scan of the brain.
As the databases of IRIS® Registry and LIFE-Adult-Study are both large and complex, our team is participating in the analysis work of these two databases using big-data-related methods. For IRIS® Registry, we investigate the association of demographic features, social histories, and onset clinical characteristics of different diseases. We also focus on the clinical stage of diseases and their relationship with other factors. For the LIFE-Adult-Study, we associate the ophthalmic imaging data (fundus imaging and OCT results) with chemical measurements to find predictive biomarkers of certain diseases, like age-related macular edema and diabetic retinopathy.
Responsible AI: Visual field predictions and analysis
Recent advancements in deep learning have focused on predicting patients’ future visual fields, offering the potential to anticipate disease progression and tailor interventions more effectively. In our research, we selected two prominent methods from the literature and replicated them to evaluate their performance on a clinical dataset. While the reported metrics in the literature were confirmed to be accurate, our findings reveal a significant bias in these models, particularly favoring stable patients.
This bias leads to unacceptable performance when it comes to identifying patients with worsening conditions, which is a critical aspect of clinical utility. The algorithms often fail to detect these more severe cases, thereby missing the very patients who would benefit most from timely interventions. Our study highlights the need for further refinement of these models to ensure they can reliably identify and prioritize worsening cases in diverse clinical settings.

Visual Field Toolbox
We are committed to open science, and in addition to sharing our AI model, we also provide tools for researchers and developers to accelerate scientific progress and benchmark implementations. To support this commitment, we have developed the first Python package, PyVisualFields, dedicated to visual field analysis. This package offers functionalities for visualization, normalization, progression analysis, and more. PyVisualFields is available for installation via PyPI, and we are continually maintaining and expanding its features in future versions.

Aging and lifestyle affecting the eye
Aging and lifestyle affect the eye and can be risk factors for ocular diseases or confounders for their diagnosis. Many lifestyle-related parameters, such as obesity, high blood pressure, unfavorable cholesterol values, the lack of physical activity, or the consumption of alcohol or tobacco, may impair retinal structure. While lifestyle-related retinal changes are often too small to be noticed by people, they may impose risk factors for severe ocular diseases, including potentially blinding optic neuropathies.
As reported, exfoliation syndrome (XFS), a degenerative age-related condition that can develop into glaucoma, was increasing with age and more frequent in women and asthma patients. And people with frequent dietary fiber-rich vegetables and fruit consumption were less likely to have XFS. Aging and many factors have been approved to increase the onset and progress risk of diabetic retinopathy, such as long duration of diabetes, hyperglycemia, hypertension, hyperlipidemia, smoking, moderate alcohol consumption, obesity, and pregnancy, etc. Similarly, identified risk factors for retinal artery occlusion include hyperglycemia, hypertension, hyperlipidemia, smoking, and advanced age.
Previous studies infer that it’s necessary to deeply and comprehensively investigate the effects of aging and lifestyle on eyes, which requires a large population-based systemic clinical cohort study. It will be much beneficial to both patients and healthcare providers to find crucial factors that lead to the development and progression of certain ocular diseases like glaucoma, age-related macular degeneration, and diabetic retinopathy, especially during the early stage. Thus, healthcare providers will be able to work out a plan to better control the diseases before the onset and retard the progression.
Our team is curious about how aging and lifestyle factors affect the eye. We are participating in a population-based study (with about 10 thousand participants) that includes ocular imaging and a large number of physiological and cognitive parameters to systematically investigate the relationship between retinal parameters obtained via fundus photography and optical coherence tomography (OCT) and age, as well as various groups of lifestyle-related variables, such as cardiovascular parameters (e.g., blood pressure, cholesterol, history of strokes), anthropometric parameters (e.g., body mass index, waist-to-hip ratio), substances (e.g., alcohol and tobacco consumption), or neurological diseases (e.g., Parkinson’s disease, neuro-cognitive disorders, multiple sclerosis). This project contributes directly and immediately to public health by exploring the relationship between ocular diseases, demographic characteristics, and lifestyle-related parameters.
Responsible AI: Fundus Image Analysis and Benchmarking
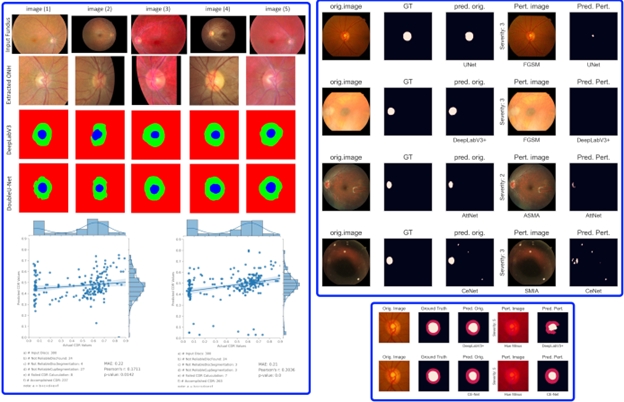
The aim of this project is to assess the impact of fundus image quality on the performance of AI-based diagnostic and assessment methods. Additionally, adversarial attacks are recognized as a threat to the deployment of AI in medicine.
To address these challenges, various image perturbation techniques are employed to simulate different conditions of fundus image capture. Some of these perturbation methods are specific to fundus images. Both white-box and black-box adversarial perturbations are also considered.
We benchmark the performance of several AI tasks, including disc and cup segmentation, as well as diabetic retinopathy and glaucoma detection. Multiple AI models are applied, and their results are analyzed. The findings indicate that image quality and adversarial perturbations have a significant impact, revealing that the robustness of AI methods varies even when using the same training datasets and scenarios. This underscores the importance of considering these perturbations before deploying AI models in clinical settings.
In another study, we focused on cup-to-disc ratio estimation and demonstrated that AI models trained on publicly available datasets are not yet ready for deployment in clinical settings. Our findings reveal significant errors when these models are applied to external, unseen datasets from other hospitals, especially when the data distribution is more diverse.

Early Identification for Pre-onset High-Risk Glaucoma Patients
Glaucoma, a leading cause of blindness, affects over 76 million people globally and over 3 million in the U.S., with many unaware of their condition. This “silent thief of sight” disproportionately impacts older adults, particularly African Americans and Hispanics, leading to significant vision loss and healthcare costs. With glaucoma prevalence expected to rise to 111.8 million by 2040, early detection and treatment are crucial for preventing irreversible vision loss and reducing the disease’s socioeconomic impact.
To address this, we aim to develop AI-powered tools for early detection and prediction in glaucoma care, focusing on primary open-angle glaucoma (POAG). These models will integrate multi-modal data, including imaging, visual fields, electronic health records, and genomics, to improve prediction accuracy. Our preliminary study using deep learning and visual field data has shown promise in identifying early glaucoma biomarkers. However, to enhance predictive accuracy, further integration of diverse data is necessary. Challenges include dealing with imbalanced datasets, varying disease onset timelines, and inconsistent data availability across different modalities.